Computational modeling yields a protein fragment that could bind to coronavirus spike proteins and destroy them.
The research described in this article has been published on a preprint server but has not yet been peer-reviewed by scientific or medical experts.
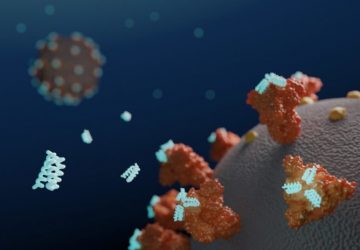
Using computational models of protein interactions, researchers at the MIT Media Lab and Center for Bits and Atoms have designed a peptide that can bind to coronavirus proteins and shuttle them into a cellular pathway that breaks them down.
This type of peptide could hold potential as a treatment that would prevent the SARS-CoV-2 virus from reproducing itself within infected cells, the researchers say.
“Our idea was to use computational techniques to engineer a peptide that could be a therapeutic for Covid-19. Once the peptide gets in the cell, it can simply tag and degrade the virus,” says Pranam Chatterjee, a recent MIT PhD recipient and the lead author of the study.
The researchers have tested the peptide in human cells, and they are now planning additional cell and animal studies to further evaluate its efficacy. They reported their initial findings in a preprint posted on bioRxiv, an online preprint server, on June 1, and have also submitted it to a peer-reviewed journal. Graduate student Manvitha Ponnapati and Joseph Jacobson, an associate professor in the MIT Media Lab, co-authored the study.
Modeling peptides
Scientists are pursuing many different strategies to develop new therapeutics against SARS-CoV-2. One area of interest is developing antibodies that bind to and inactivate viral proteins such as the spike protein, which coronaviruses use to enter human cells. A related approach uses small protein fragments called peptides instead of antibodies.
The MIT team set out to engineer peptides that could strongly bind to the spike protein inside cells, and to use these peptides to trigger the cells to break down the viral proteins. Their idea was to have their peptides recruit naturally occurring proteins called E3 ubiquitin ligases, which can mark proteins for destruction when cells no longer need them.
To generate their spike-protein-binding peptides, the researchers used a computational model of protein interactions that they had previously trained to optimize binding strength between two proteins. Chatterjee and others recently used similar computational methods to design improved versions of enzymes used for the genome-editing technique known as CRISPR. Their new CRISPR-Cas9 enzymes, together, can target more than 70 percent of DNA sequences, while the most commonly used form of CRISPR-Cas9 reaches only about 10 percent.
In this case, the researchers used as their starting point the human ACE2 protein, which is found on the surface of certain types of human cells and binds to the coronavirus spike protein.
They used their model to break ACE2 into many small fragments and then computationally predict how the fragments would interact with the spike protein. They instructed the model to optimize three features: First, they engineered peptides to have strong binding affinity to the spike protein. Second, they established that the peptides could bind well to other coronavirus spike proteins, in hopes that it could work against past and future strains of coronaviruses. Third, they ensured that the peptides would not bind strongly to human proteins called integrins, which are the proteins that normally bind to the ACE2 receptor in the body.
This process generated about 25 candidate peptides, which the researchers fused to an E3 ubiquitin ligase and tested in human cells that expressed a fragment of the spike protein known as the receptor-binding domain (RBD).
The best of these candidates, a 23-amino-acid peptide, broke down about 20 percent of the RBD proteins in the cells. However, this peptide did not work as well as the original ACE2 protein, which broke down about 30 percent of the RBD proteins. To improve the peptide’s performance, the researchers used their model to simulate how its RBD-binding would be affected if they substituted different amino acids at each of its 23 positions. That optimization process yielded a mutant peptide that improved the degradation rate to over 50 percent.
Tagged for destruction
One key advantage of this peptide is its small size — even when fused to the E3 ubiquitin ligase, the entire chain is only around 200 amino acids in length. The researchers envision that RNA or DNA encoding the peptides could be delivered by harmless viruses called adeno-associated viruses.
Another possibility would be to deliver the peptide on its own, allowing it to bind to the coronavirus spike protein outside of cells and be carried into cells with the virus. In that case, the virus would then be tagged for destruction as soon as it enters the cell, Chatterjee says.
The researchers are now planning to test the peptide in human cells infected with the SARS-CoV-2 virus, which will occur at specialized biosafety labs outside MIT. If those tests are successful, the researchers hope to test the peptide in animal models. They are also working on further improving the peptide so that it can bind the spike protein more strongly.
This work was supported by the consortium of sponsors of the MIT Media Lab, the MIT Center for Bits and Atoms, and Jeremy and Joyce Wertheimer.
source: www.scitechdaily.com